ميگاڤيروس
| ميگاڤيروس Megavirus | |
|---|---|
| |
| تصنيف الفيروسات | |
| Group: | Group I (dsDNA)
|
| (unranked): | |
| Family: | Megaviridae
|
| Genus: | Megavirus
|
| Species | |
|
Megavirus chilensis | |
ميگاڤيروس Megavirus[1] هو جنس من الڤيروسات يندرج تحته نوع واحد معروف يسمى ڤيروس چيلنسيس (MGVC)، مرتبط تطوريا مع أكانثاموبا پوليفاگا ميميڤيروس (APMV).[2]
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
الاكتشاف
تم عزل ميگاڤيروس من عينة مياه في أبريل 2010 أخذت من سواحل تشيلي، بالقرب من محطة بحرية في لاس كروكس، بواسطة البروفيسور جان-مايكل كلاڤري ود. چانتال أبرگيل من معمل المعلومات الجينية والبنيوية بجامعة أيكس-مارسيليا]].
التصنيف
Megavirus is not yet classified by the International Committee on Taxonomy of Viruses, but will be proposed as a member of the Megaviridae, a new family constituted of the large DNA viruses the genome of which is around a million base pairs in length. Members of this new family defined by a number of common specific features (Table 1 and 2) will include various viruses likely to share a common ancestor with Mimivirus and Megavirus, although their present genome size was reduced below 1 Mb. Megavirus also joins a group of large viruses known as nucleocytoplasmic large DNA viruses (NCLDV), although this term appears increasingly inappropriate to designate viruses replicating entirely within the cytoplasm of their hosts through the de novo synthesis of large virion factories. Megavirus and Mimivirus share 594 orthologous genes, mostly located within the center segment of their genomes. At the amino-acid sequence level, the corresponding proteins share an average of 50% identical residues.[بحاجة لمصدر]
التركيب
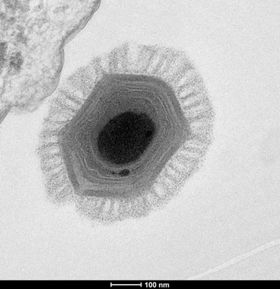
يتكون الميجا فيرس من قفيصة بروتينية قطرها 440 نانومتر ، محاطة بكبسولة يتراوح سمكها بين 75-100 نانومتر
الجينوم
جينوم الميجا فيروس هو جزيء خطي ذو طاقين من الحمض النووي الريبوزي منقوص الأوكسجين ، يمتلك 1,259,197 زوج من القواعد النتيروجينية. وبالتالي يصبح هو اكبر جينوم فيروسي حتى الآن حيث يتفوق على ثاني اكبر جينوم فيروسي الا وهو ماما فيروس ب 67,500 زوج من القواعد.
The Megavirus chilensis genome is a linear, double-stranded molecule of DNA with 1,259,197 base pairs in length. This makes it the largest viral genome deciphered so far, outstripping the next-largest virus genome of Mamavirus by 67.5 kb. Prior validation of its transcriptome, it is predicted to encode 1,120 protein-coding genes, a number largely above the one exhibited by many bacteria.[بحاجة لمصدر]
However, beyond an incremental change in size, the Megavirus genome exhibits 7 aminoacyl tRNA synthetases (Table 2), the archetypes of enzymes previously thought only to be encoded by cellular organisms. While 4 of these enzymes were already present in Mimivirus and Mamavirus (for tyrosine, arginine, cysteine, and methionine), Megavirus is exhibiting three more 3 (for tryptophane, asparagine, and isoleucine). Interestingly, the unique aminoacyltRNA synthetase encoded by Cafeteria roenbergensis virus corresponds to the one for isoleucine. Megavirus also encodes a fused version of the mismatch DNA repair enzyme MutS, uniquely similar to the one found in the mitochondrion of octocorals. This puzzling MutS version appears to be a trademark of the Megaviridae family.[3] Like Mimivirus and CroV, Megavirus contains many genes for sugar, lipid and amino acid processing, as well as some metabolic genes not found in any other virus.[4]
النسخ المتماثل
Megavirus replication stages closely follows the one already described for Mimivirus. Following rapid engulfing by phagocytosis, and the delivery of the particle core to the cytoplasm, begins the eclipse phase. A closer examination indicates the presence of cytoplasmic “seeds”, of sizes comparable to the most internal membrane-enclosed core of the Megavirus particle. These seeds then develop in full bloomed virion factory over the following 14 hours. The full course of infection (until the complete lysis of the amoebal cells) takes 17h in average, compared to 12 h for Mimivirus. The average number of released Megavirus particles (i.e. the “burst size”) is about half the thousand produced by Mimivirus.[بحاجة لمصدر]
As expected from a virus encoding its own complete DNA replication, repair, and transcription machinery, the host nucleus does not appear to be involved in any of Megavirus replication stages, as already seen for Mimivirus.[بحاجة لمصدر]
تأثيرات الاكتشاف
Two Megavirus specific features are of fundamental importance. First, even if its new record-sized genome only represent a small 6% increment compared to the second largest Mamavirus genome, it indicates that we have not yet reached the limit of viral genome sizes, and that even more complex viruses may remain to be discovered,[بحاجة لمصدر] most likely in aquatic environment, the viral diversity of which has been barely scratched by recent metagenomic studies.[بحاجة لمصدر]
Second, the presence of three additional aminoacyltRNA synthetases in the Megavirus genome, with a total of seven, definitely rules out their independent acquisition by lateral transfer from a host.[بحاجة لمصدر] In addition, the phylogenic analysis of their amino acid sequences does not cluster them with any known protozoan clade, but rather connect them deeply to eukaryotic domain.[بحاجة لمصدر]
The conclusion then becomes inescapable that the genome of these giant viruses originated from an ancestral cellular genome (thus endowed of a translation apparatus, with all 20 aminoacyl tRNA synthetases) from which today’s Megaviridae derived by a number of lineage specific genome reduction events, a scenario akin to the one followed by all parasitic organisms. As indicated by the deep rooting of their phylogeny, the Megaviridae lineage might be very old, eventually predating the emergence of modern eukaryotes, or be contemporary and/or linked to the emergence of the nucleus. 5 Alternatively, it could be derived from an extinct cellular domain (the controversial 4th domain of the Tree of Life[5]), many genes of which would have managed to persist in today’s giant viral genomes.[بحاجة لمصدر]
جدول1 : أكبر الڤيروسات العملاقة مع تسلسل الجينوم الكامل
| اسم الڤيروس العملاق | طول الجينوم | الجينات | Capsid diameter (nm) | Hair cover | Genbank # |
|---|---|---|---|---|---|
| Megavirus chilensis[1] | 1,259,197 | 1120 proteins (predicted) | 440 | yes (75nm) | JN258408 |
| Mamavirus[6] | 1,191,693 | 1023 proteins (predicted) | 390 | Yes (120 nm) | JF801956 |
| Mimivirus[2][7] | 1,181,549 | 979 proteins 39 non-coding | 390 | Yes (120 nm) | NC_014649 |
| M4[8] (Mimivirus « bald » variant) | 981,813 | 756 proteins (predicted) | 390 | No | JN036606 |
| Cafeteria roenbergensis virus[9] | 617,453 (730 kb) | 544 proteins (predicted) | 300 | No | NC_014637 |
جدول 2: الخصائص الشائعة في الڤيروسات العملاقة
| اسم الڤيروس العملاق | Aminoacyl-tRNA synthetase | Octocoral-like MutS | Stargate[10] | Known virophage[11] | Cytoplasmic virion factory | Host |
|---|---|---|---|---|---|---|
| Megavirus chilensis | 7 (Tyr, Arg, Met, Cys, Trp, Asn, Ile) | yes | yes | no | yes | Acanthamoeba (Unikonta, Amoebozoa) |
| Mamavirus | 4 (Tyr, Arg, Met, Cys) | yes | yes | yes | yes | Acanthamoeba (Unikonta, Amoebozoa) |
| Mimivirus | 4 (Tyr, Arg, Met, Cys) | yes | yes | yes | yes | Acanthamoeba (Unikonta, Amoebozoa) |
| M4 (Mimivirus « bald » variant) | 2 (Met, Cys) | no | yes | resistant | yes | Acanthamoeba (Unikonta, Amoebozoa) |
| Cafeteria roenbergensis virus | 1 (Ile) | yes | no | yes | yes | Phagotrophic protozoan (Heterokonta, Stramenopiles) |
انظر أيضا
- ميميڤيروس
- Cafeteria roenbergensis virus - the largest marine virus
- Papilloma virus – smallest known single stranded DNA viruses
المصادر
- ^ أ ب Arslan, D.; Legendre, M.; Seltzer, V.; Abergel, C.; Claverie, J.-M. (2011). "Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae". Proceedings of the National Academy of Sciences. doi:10.1073/pnas.1110889108.
- ^ أ ب Raoult, D.; Audic, S; Robert, C; Abergel, C; Renesto, P; Ogata, H; La Scola, B; Suzan, M; Claverie, JM (2004). "The 1.2-Megabase Genome Sequence of Mimivirus". Science. 306 (5700): 1344–50. doi:10.1126/science.1101485. PMID 15486256.
- ^ Ogata, Hiroyuki; Ray, Jessica; Toyoda, Kensuke; Sandaa, Ruth-Anne; Nagasaki, Keizo; Bratbak, Gunnar; Claverie, Jean-Michel (2011). "Two new subfamilies of DNA mismatch repair proteins (MutS) specifically abundant in the marine environment". The ISME Journal. 5 (7): 1143–51. doi:10.1038/ismej.2010.210. PMC 3146287. PMID 21248859.
- ^ Claverie, Jean-Michel; Abergel, Chantal (2010). "Mimivirus: The emerging paradox of quasi-autonomous viruses". Trends in Genetics. 26 (10): 431–7. doi:10.1016/j.tig.2010.07.003. PMID 20696492.
- ^ Dickey Zakaib, Gwyneth (2011). "The challenge of microbial diversity: Out on a limb". Nature. 476 (7358): 20–1. doi:10.1038/476020a. PMID 21814255.
- ^ Colson, P.; Yutin, N.; Shabalina, S. A.; Robert, C.; Fournous, G.; La Scola, B.; Raoult, D.; Koonin, E. V. (2011). "Viruses with More Than 1,000 Genes: Mamavirus, a New Acanthamoeba polyphaga mimivirus Strain, and Reannotation of Mimivirus Genes". Genome Biology and Evolution. 3: 737–42. doi:10.1093/gbe/evr048. PMC 3163472. PMID 21705471.
- ^ Legendre, Matthieu; Santini, Sébastien; Rico, Alain; Abergel, Chantal; Claverie, Jean-Michel (2011). "Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing". Virology Journal. 8: 99. doi:10.1186/1743-422X-8-99. PMC 3058096. PMID 21375749.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Boyer, M.; Azza, S.; Barrassi, L.; Klose, T.; Campocasso, A.; Pagnier, I.; Fournous, G.; Borg, A.; Robert, C. (2011). "Mimivirus shows dramatic genome reduction after intraamoebal culture". Proceedings of the National Academy of Sciences. 108 (25): 10296–301. doi:10.1073/pnas.1101118108. PMC 3121840.
- ^ Fischer, M. G.; Allen, M. J.; Wilson, W. H.; Suttle, C. A. (2010). "Giant virus with a remarkable complement of genes infects marine zooplankton". Proceedings of the National Academy of Sciences. 107 (45): 19508–13. doi:10.1073/pnas.1007615107. PMC 2984142.
- ^ Zauberman, Nathan; Mutsafi, Yael; Halevy, Daniel Ben; Shimoni, Eyal; Klein, Eugenia; Xiao, Chuan; Sun, Siyang; Minsky, Abraham (2008). Sugden, Bill (ed.). "Distinct DNA Exit and Packaging Portals in the Virus Acanthamoeba polyphaga mimivirus". PLoS Biology. 6 (5): e114. doi:10.1371/journal.pbio.0060114. PMC 2430901. PMID 18479185.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Fischer, M. G.; Suttle, C. A. (2011). "A Virophage at the Origin of Large DNA Transposons". Science. 332 (6026): 231–4. doi:10.1126/science.1199412. PMID 21385722.
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
قراءات إضافية
- Ghedin, Elodie; Claverie, Jean-Michel (2005). "Mimivirus relatives in the Sargasso sea". Virology Journal. 2: 62. doi:10.1186/1743-422X-2-62. PMC 1215527. PMID 16105173.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - Monier, Adam; Claverie, Jean-Michel; Ogata, Hiroyuki (2008). "Taxonomic distribution of large DNA viruses in the sea". Genome Biology. 9 (7): R106. doi:10.1186/gb-2008-9-7-r106. PMC 2530865. PMID 18598358.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
وصلات خارجية
- GiantVirus.org – an information resource on the genome of giant viruses.
- International Committee on Taxonomy of Viruses (ICTV) picture gallery - images of mimivirus
- Van Etten, James L. (2011). "Giant Viruses". American Scientist. 99 (4): 304. doi:10.1511/2011.91.304.